Genes, Brains, and Intelligence: What’s New?
Emil O. W. Kirkegaard, American Renaissance, March 2, 2021

In the second half of the 20th century, Marxist intellectuals such as Stephen Jay Gould and Richard Lewontin stigmatized research into the study of intelligence, especially of individual and group differences. Although still a taboo topic, over the last few years there has been a constant stream of new research on genetics and intelligence, so much so that most people have a hard time keeping up. This is all the more impressive in light of the cultural revolution on college campuses and the revival of blank slate ideology by cultural elites.
One can attribute much of the progress of research on the genetics of intelligence to the decades-long work of Robert Plomin and Ian Deary. This power duo (both head prominent research labs) steered clear of ethnic and racial differences research, resolutely focusing on individual differences. The result was the establishment of increasingly large datasets, many of which include family members, especially twins (TEDS is an example). These have in turn produced a torrent of findings showing the powerful effects of genetics on human psychology and life outcomes ranging from educational attainment to marital stability.
Ian Deary (Credit Image: CCACE / The University of Edinburgh)
Ian Deary and two colleagues, Simon R. Cox and W. David Hill (all of University of Edinburgh), have a review article in Molecular Psychiatry. This is a prestigious journal. It may surprise some readers that journal prestige is unrelated to scientific rigor. It does, however, work as an indicator of how much social status academics award to a topic. It’s now almost impossible to find prestigious outlets publishing the genetic-denialist writings that were typical some decades ago. The abstract of this paper pulls no punches:
Individual differences in human intelligence, as assessed using cognitive test scores, have a well-replicated, hierarchical phenotypic covariance structure. They are substantially stable across the life course, and are predictive of educational, social, and health outcomes. From this solid phenotypic foundation and importance for life, comes an interest in the environmental, social, and genetic aetiologies of intelligence, and in the foundations of intelligence differences in brain structure and functioning. Here, we summarise and critique the last 10 years or so of molecular genetic (DNA-based) research on intelligence, including the discovery of genetic loci associated with intelligence, DNA-based heritability, and intelligence’s genetic correlations with other traits. We summarise new brain imaging-intelligence findings, including whole-brain associations and grey and white matter associations. We summarise regional brain imaging associations with intelligence and interpret these with respect to theoretical accounts. We address research that combines genetics and brain imaging in studying intelligence differences. There are new, though modest, associations in all these areas, and mechanistic accounts are lacking. We attempt to identify growing points that might contribute toward a more integrated ‘systems biology’ account of some of the between-individual differences in intelligence.
The reader who has been following the field may ask, “What’s new?” In a way, not much. Many of the fundamental findings have not changed since Barbara Burks (and others) pioneered behavioral genetics in the 1920s. This is not a bad thing. Robust science is expected to produce consistent findings across many decades. At the same time, there is a lot that is new on the details of genetic causation and neuroscience.
To begin with the first, large-scale collaborations have enabled pooling of datasets to investigate the genetic basis of human traits, including psychological traits. These studies are called GWAS (genome-wide association studies). GWASs are conducted by searching across the entire human genome for genetic variations that relate to the trait of interest. This contrasts with looking only for pre-selected regions thought to be important, as was common from 2000 to 2012. This approach was a complete failure.
Credit Image: © Bobylev Sergei/ITAR-TASS/ZUMAPRESS.com
As it turns out, the human genome is huge, with some 3.1 billion base pairs. Most of these, however, don’t vary between individuals much or at all (that’s why it is said that humans are all 99 percent or more genetically identical). A small proportion of loci (locations) in the genome do vary appreciably between individuals and ancestral groups (ethnicities/races/clusters, call them what you will). There are a few million loci in which there is at least a 1 percent frequency of variation in some population. These common variants are the focus of most GWASs.
The typical GWAS involves genotyping a large number of people (usually from a saliva sample), and then measuring or recording their traits, such as height or educational attainment. As it turns out, 10,000 people is not a large enough sample to find out much of interest, because of the large number of genetic variants typically associated with a trait (this is known as polygenicity). So, to go further, researchers pool the results from different datasets and average them. The result is a meta-analysis of how much each genetic variant (called SNPs, pronounced “snips”) is related to traits of interest. The SNPs that show a relationship far beyond chance levels are called genome-wide significant hits, or “hits” for short.
Once hits are identified, it is possible to begin figuring out which genes are related to which traits. A gene may span between 1,000 or even 2 million base pairs in the genome. Most, however, are short, stretching for maybe 5,000 base pairs. Upon finding that a given genetic variant is related to some trait, one can look up a map of the genome and see which genes are close by. Often one finds that no gene is close by, raising questions of why that genetic variant is related to the trait. Perhaps it is a chance occurrence, or due to long-distance correlations to causal variants, or reflects undiscovered genetic functionality. Some of these genes have functions that are only partly understood.
Scientists figure out such functions by looking where in the body the genes are expressed (i.e., where the proteins they code for are produced). A gene that is expressed in the brain and few other places is thought to have some brain-related function. Maybe it builds neurons, or repairs them, or removes toxic chemicals from the brain. It is not always easy to find out what they do, but we can at least know that a genetic sequence is active in the brain.

Credit Image: © Cultura via ZUMA Press
What have these new scientific methods found? Where are the genetic variants responsible for intelligence? Not surprisingly, genes influence intelligence by affecting how our brains are built and how they function. As the authors put it, “Genetic variants associated with intelligence test scores tend to cluster in groups of genes linked with neurogenesis, the synapse, neuron differentiation, and oligodendrocyte differentiation.”
There are various ways to use genetic data to estimate heritability (the proportion of observed variation that is due to genetics). Each method has limitations, but they all find that intelligence is highly heritable. Still, the estimates produced by genetic analysis do not reach the same high figures as those from family studies, such as those that estimate the heritability of intelligence by studying identical twins separated at birth and reared apart. This is the so-called missing heritability problem.
There are many reasons for the missing heritability. One obvious reason is that current studies do not measure all the genetic variants — they ignore rare and complex variants. If intelligence is substantially about avoiding mutations that decrease functionality, then we will have to measure these rare variants to capture the genetic effects. Still, we can now be confident that there is a lot of genetic influence and that decades of family studies have yielded correct results.
We can go further. By finding the relationships between specific genetic variants and observed traits, we can predict phenotypic status, such as height or likelihood of having ADHD. This is done essentially by taking a weighted average of the estimated relationships of variants across the genome to the trait in question. These weighted averages are called polygenic scores. From a theoretical perspective, polygenic scores are an approximation of the genetic potential of an individual for that trait. Current polygenic scores are not perfect — they do not capture all the genetic variation. Still, they are reaching levels that are useful for research and in some cases, for clinical applications (mostly in medicine).
The strength of these polygenic scores is a function of many factors, including the complexity of the genetic architecture (simpler architecture is easier to understand), how difficult the trait is to measure (more measurement error is worse), the number of people we can recruit and genotype, the quality of the genotyping, and the algorithms used on the data. Progress is being made in all these areas, some of it quite rapid.

Credit Image: © Image Source/ZUMAPRESS.com
There are not one, but three recent GWASs on intelligence: One used data from almost 250,000 people (led by Hill, coauthor of the new review), one by Davies et al. used about 300,000, and one by Savage et al. used about 270,000. Each study found a long list of variants related to intelligence, mostly replicating each other’s findings (replication indicates good science). There was also a GWAS for educational attainment that had more than 1 million subjects.
How good are the predictions? At present, measured intelligence correlates at about 0.3 with genetic scores (that is about 10 percent of the variance). This is larger than the correlation between family income and children’s intelligence scores, so, while not clinically useful, the value is nothing to scoff at. Unfortunately, studies using siblings find that about half of this validity seems to be confounded by family factors, since it does not work between siblings. Thus, there is a need for larger GWAS samples and, preferably, samples of siblings and other family members, so that one can adjust for this.
Genetic tests cannot yet take the place of pencil-and-paper tests, but as polygenic scores improve, they are bound to replace testing. In the future, everybody will have his genome data available from birth, and can simply consent to share it with interested parties for predictive purposes.
Another set of findings from genetic studies is that one can now compute genetic correlations between traits. Essentially, this estimates the genetic overlap between traits due to gene variants that influence more than one trait (pleiotropy), or that are close together in the genome and thus tend to occur together. Again, the research finds that the genetic overlap mirrors the relationships we see in the real world. Thus, for example, the negative association between intelligence and ADHD is not due entirely to socialization, but also results from shared genetic causes. Similarly, research finds that smarter people have better physical health, and this relationship is substantially due to genetic overlap.
Traditional family studies had reached similar conclusions. There is even a study showing that this mirroring of phenotypic and genetic associations is a general feature; it is known as Cheverud’s Conjecture. Thus, whenever you hear of some association, e.g. homosexuality and mental illness, you have good justification for assuming a partial genetic basis for it (this specific example was confirmed by a recent GWAS, Figure 4). Section C in the figure below illustrates associations of various traits with intelligence.
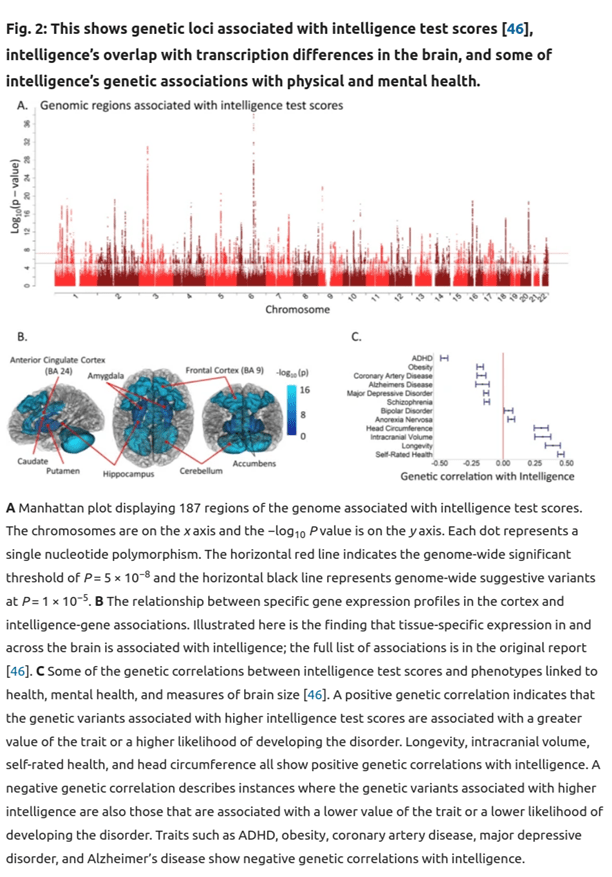
Click here for an enlarged version.
These findings are about associations between traits in individuals, not ancestry group averages, but if one were to extrapolate within reason, one conclusion could be that group — or racial — phenotypic differences reflect genetic ones. Following Arthur Jensen, we might call that the default hypothesis.
What’s new in development of brains? There are a host of recent studies finding that larger brains tend to be better. The authors summarize this finding:
A meta-analysis of data from over 148 studies across more than 8000 individuals [71] estimated the association at r = 0.24. A re-analysis of those data including only healthy adults estimated the association at r = 0.31; this rose to r = 0.39 when it included only the studies judged to have used better-quality intelligence testing [72]. In a single sample of 18,426 middle- and older-aged participants of the UK Biobank (age range 44–81 years), the association between intelligence and total brain volume was estimated at r = 0.276 (95% CI = 0.252, 0.300) [73]. This is about halfway between the two previous estimates, and has the benefit of eliminating cross-cohort heterogeneity that can influence meta-analytic results.
Despite many lies and obfuscations, mainstream science is increasingly siding with the hereditarians on many of their central claims. However, as with the polygenic scores, a correlation of brain size with intelligence of about .30 gets you only so far. The rest of the Deary paper reports the associations of many other brain features with intelligence.
The human brain is the most complex organ; it is not surprising that it is hard to figure out how it works. Still, progress is being made in much the same way as with GWAS studies, thanks to more and better data. Every year, large new datasets are published, thus enabling rapid progress.
Image by Gordon Johnson from Pixabay
One team of researchers may look at finer volumes of brain structures, another may look at connections between these, and still another may look at signs of brain damage. Each of these may explain part of the puzzle, and by combining them, we should eventually be able to predict intelligence in people from brain data better than by giving them an intelligence test.
About one study using such a combination of measures, the authors write, “A study that included brain cortical characteristics (volume, area, and thickness), total volume of subcortical structures, and measures of white matter macro- and micro-structure found that, together, they accounted for up to 18 percent of the variance in general intelligence in 73-year-olds.” There was weaker validity for younger people, perhaps because the measures of brain damage have little to work with in someone who has not yet accumulated age-related defects.
The authors end their review by looking at studies that combine three topics: genes, brains, and intelligence. Such studies find the expected results, that the genetic variants that code for brain structures also code for intelligence to some degree. This supports biological causes for the relationship. Predictions of intelligence from polygenic scores also predict brain size differences, and indeed, these appear to mediate the relationship as we would expect. Ultimately, being a smarter person is largely about having genetic variants that code for proteins that create a better functioning brain; that and good luck. The faster we can figure out these genetic variants, the sooner we can prevent the genetic deterioration of humanity, and the faster we can begin its genetic improvement.















